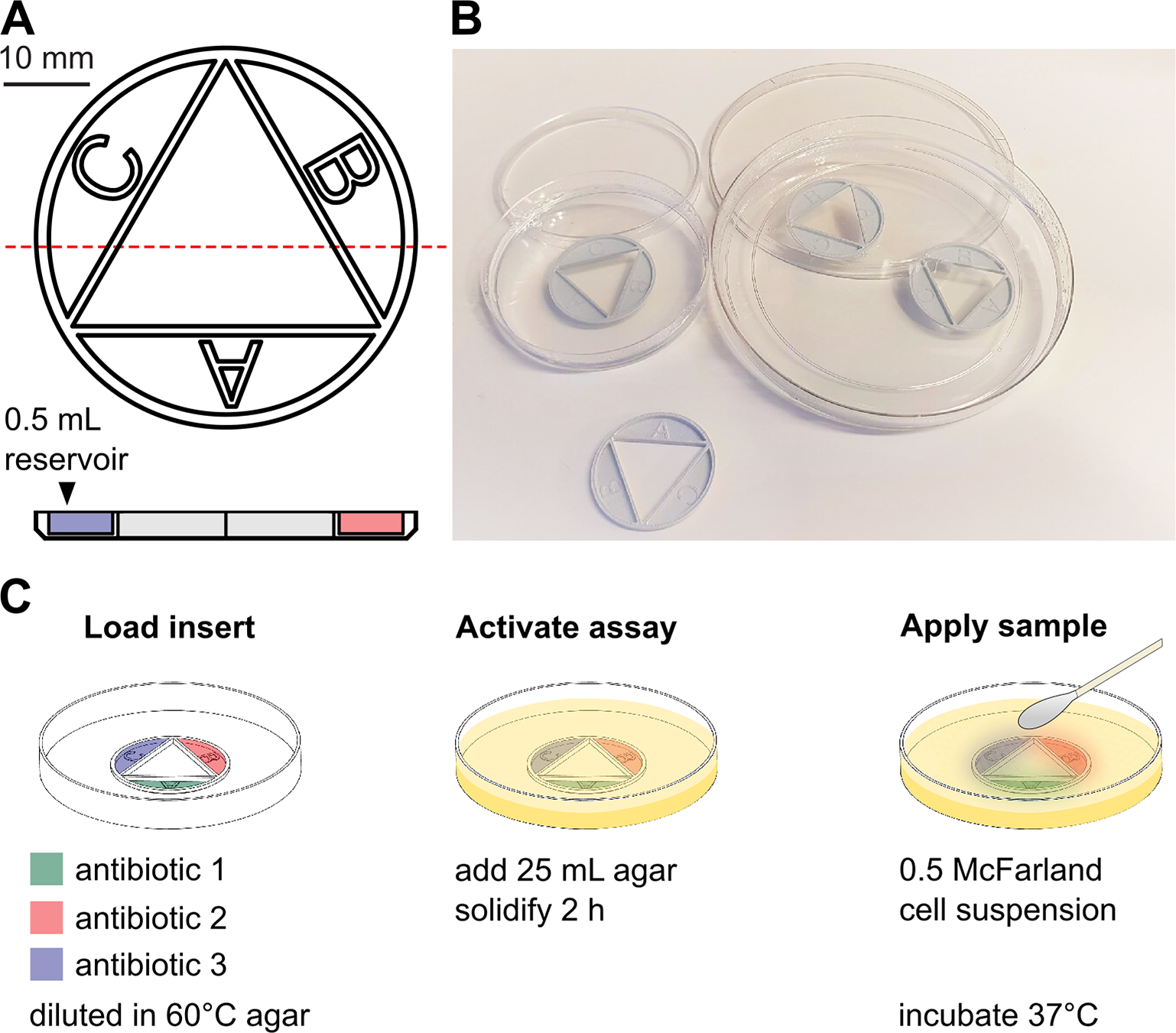
Antibiotic combination therapies are important for the efficient treatment of many types of infections, including those caused by antibiotic-resistant pathogens. Combination treatment strategies are typically used under the assumption that synergies are conserved across species and strains, even though recent results show that the combined treatment effect is determined by specific drug–strain interactions that can vary extensively and unpredictably, both between and within bacterial species. To address this problem, we present a new method in which antibiotic synergy is rapidly quantified on a case-by-case basis, allowing for improved combination therapy. The novel CombiANT methodology consists of a 3D-printed agar plate insert that produces defined diffusion landscapes of 3 antibiotics, permitting synergy quantification between all 3 antibiotic pairs with a single test.
Automated image analysis yields fractional inhibitory concentration indices (FICis) with high accuracy and precision. A technical validation with 3 major pathogens, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus, showed equivalent performance to checkerboard methodology, with the advantage of strongly reduced assay complexity and costs for CombiANT. A synergy screening of 10 antibiotic combinations for 12 E. coli urinary tract infection (UTI) clinical isolates illustrates the need for refined combination treatment strategies. For example, combinations of trimethoprim (TMP) + nitrofurantoin (NIT) and TMP + mecillinam (MEC) showed synergy, but only for certain individual isolates, whereas MEC + NIT combinations showed antagonistic interactions across all tested strains. These data suggest that the CombiANT methodology could allow personalized clinical synergy testing and large-scale screening. We anticipate that CombiANT will greatly facilitate clinical and basic research of antibiotic synergy.